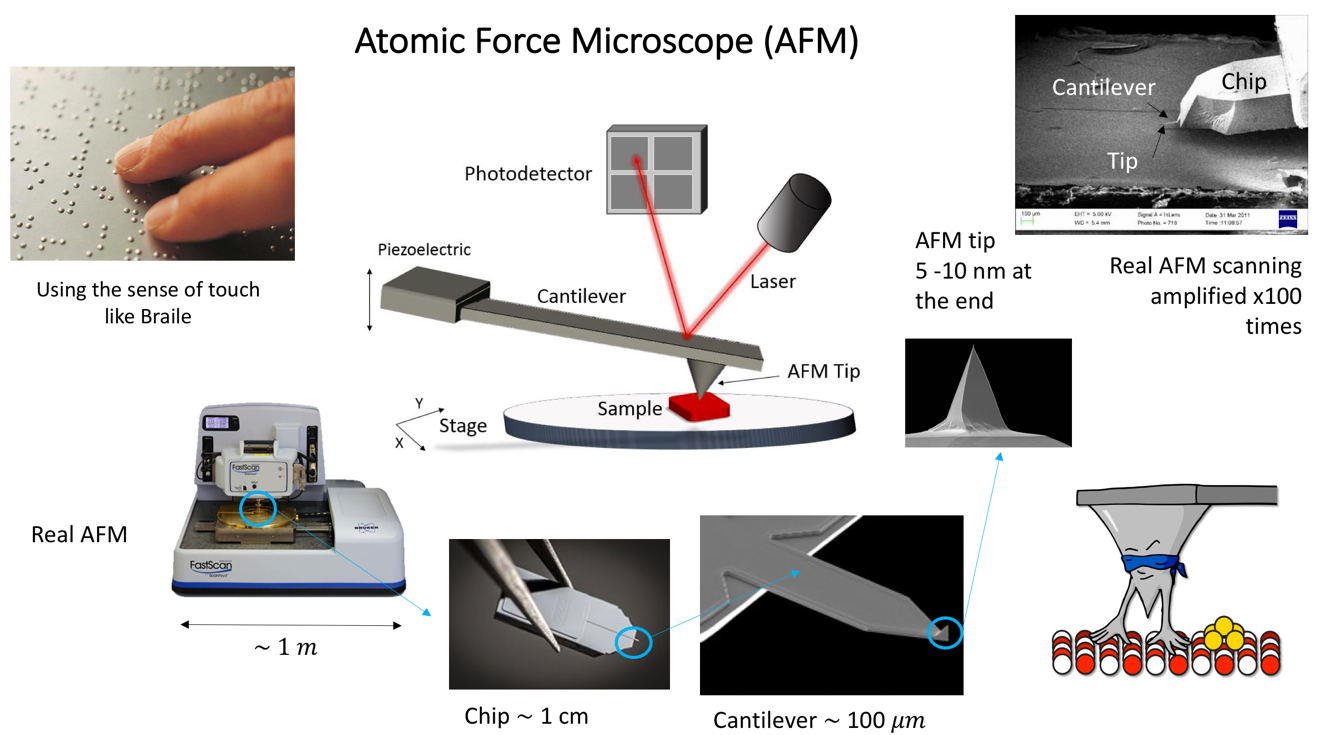
As humans, we ‘see’ because light interacting with the surfaces around us reaches our eyes. But how do we ‘see’ when want to study objects that are so small light steers around them? - like the study of objects on a nanometric scale such as individual proteins or molecules, far below the resolution limit imposed by the properties of light?
One solution to this is to use an object to 'touch' or 'feel' a surface to detect what shape it has. Basically, we ‘do Braille’!
One solution to this is to use an object to 'touch' or 'feel' a surface to detect what shape it has. Basically, we ‘do Braille’!
Like Braille, the next logical sense to be used when the sense of sight is missing is the sense of touch. Unlike any other microscope, the Atomic Force Microscopy (AFM) was invented in 1980’s with this idea that if we have a ´finger´ small enough, we could be able to touch and study soft biological objects such as bacteria cells, tissues, proteins, nucleic acids, etc.
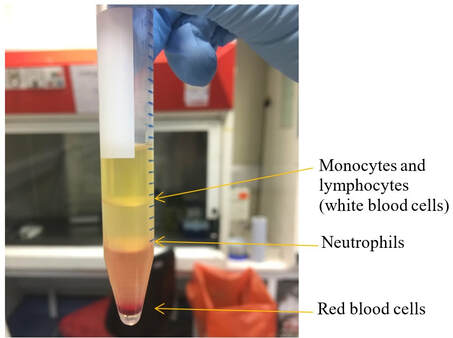
The main advantage of this technique is that we use a nanometric tip to scan entire surfaces and obtain topography of the samples without transferring energy to the sample and hence performing no harm to biological samples. At the Florey Institute we collaborate among the Departments of Physics and Astornomy, MBB and Medical School at the Hobbs’ Lab to apply AFM to study DNA interactions with other proteins and the topography of Bacteria cells.
The main advantage of this technique is that we use a nanometric tip to scan entire surfaces and obtain topography of the samples without transferring energy to the sample and hence performing no harm to biological samples. At the Florey Institute we collaborate among the Departments of Physics and Astornomy, MBB and Medical School at the Hobbs’ Lab to apply AFM to study DNA interactions with other proteins and the topography of Bacteria cells.
The landscape of the Bacteria Cell:
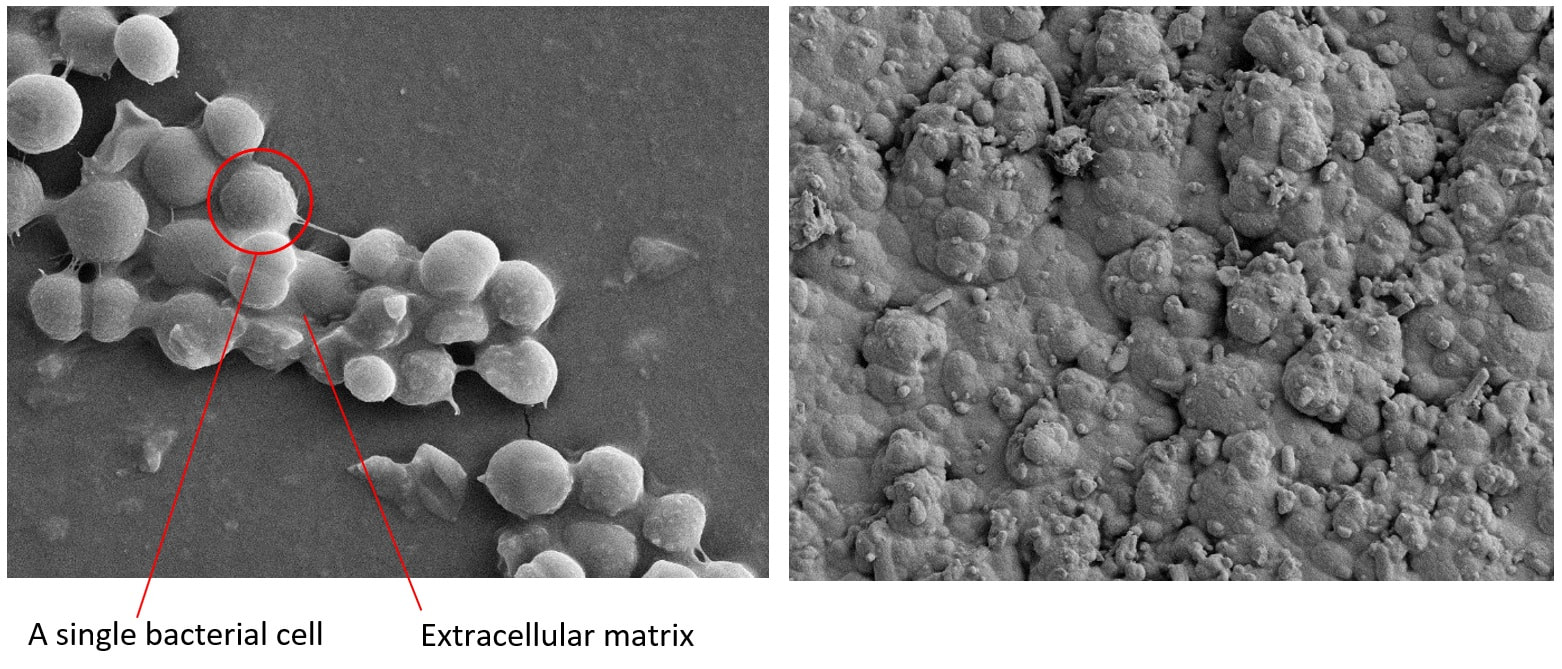
In the Hobbs’ group we are very interdisciplinary. That is why there has been a long-term collaboration between Prof. Jamie Hobbs from Physics and Prof. Simon Foster from Microbiology. Carrying out this project Laia Pasquina Lemonche, a third year PhD Cohort student is using the Atomic Force Microscope to study more in depth the topography of bacteria and the molecular organization of the cell wall from Staphylococcus aureus and Bacillus subtilis.
One of the main advantages of using this technique instead of other high resolution approaches is that we can obtain very detailed information from a single cell without the need for averaging any data. As a result, we can start answering fundamental questions about the morphology of bacteria and their behaviour but also new questions are arising from these findings. Through this, we are one step closer to better understand the host-pathogen interaction and how to treat bacterial infections effectively.
The landscape of the Bacteria Cell:
In the Hobbs’ group we are very interdisciplinary. That is why there has been a long-term collaboration between Prof. Jamie Hobbs from Physics and Prof. Simon Foster from Microbiology. Carrying out this project Laia Pasquina Lemonche, a third year PhD Cohort student is using the Atomic Force Microscope to study more in depth the topography of bacteria and the molecular organization of the cell wall from Staphylococcus aureus and Bacillus subtilis.
One of the main advantages of using this technique instead of other high resolution approaches is that we can obtain very detailed information from a single cell without the need for averaging any data. As a result, we can start answering fundamental questions about the morphology of bacteria and their behaviour but also new questions are arising from these findings. Through this, we are one step closer to better understand the host-pathogen interaction and how to treat bacterial infections effectively.
On the left - bacteria cell alive in 3D made by the AFM at 'low resolution'; On the right - 'high resolution' image of purified cell wall made by the AFM revealing the surface structures of the cell.
DNA-protein interactions:
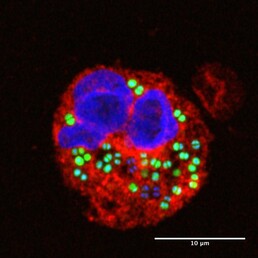
Another great application of AFM besides studying the topography of a sample, it is its ability to ‘video record’ biological reaction. For instance, DNA Polymerase, an enzyme responsible for DNA replication and repair during cell division, can be allowed to act on DNA, and consecutive AFM images of the process can be taken so as to ‘see’ the process actually happening in an environment mimicking the native cell conditions. In our lab, we are collaborating with Jon R. Sayers to use the AFM approach to understand the interaction between a DNA molecule and the Flap-endonuclease protein. Vinny Verma, a second year PhD Cohort student is using High-speed AFM to investigate this interaction to understand the conformation changes that occur during the process and how the cell manipulates the DNA using biomolecular machines.
For AFM the prospects are many, as we’re yet to see what new advancements AFM can do for 'high resolution' imaging and even what video-AFM can do for our understanding of cells!
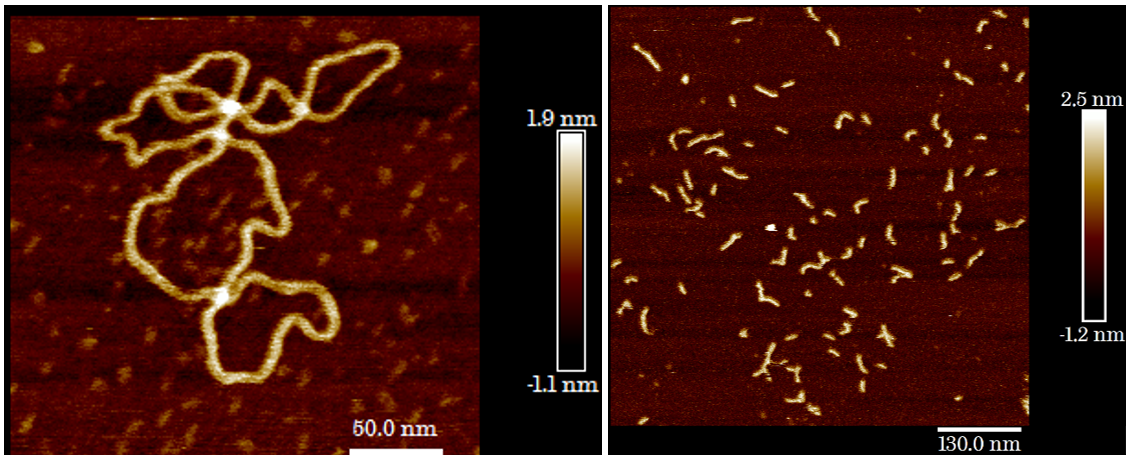
| Plasmid DNA imaged in air using AFM | Short DNA strands imaged in buffer using AFM |