Experimental approaches for studying bacteria have changed dramatically over the last 20 years. Among the most influential shifts in technology has been the increasing use of large sequencing datasets in research practice, which have allowed scientist to discover complex patterns of bacterial behaviour and reach a deeper understanding of the bacterial cell. This rapid advancement has many conceptual benefits but also comes at a significant cost, as laboratories struggle to integrate modern bioinformatics approaches with traditional microbiological research practice. But Don’t Panic. In our review article, we discuss these issues in a broad sense providing an introducing into next-generation sequencing methods for non-specialists whilst at the same time helping data-specialists understand how ‘wet’ scientists might view experimental proofs.
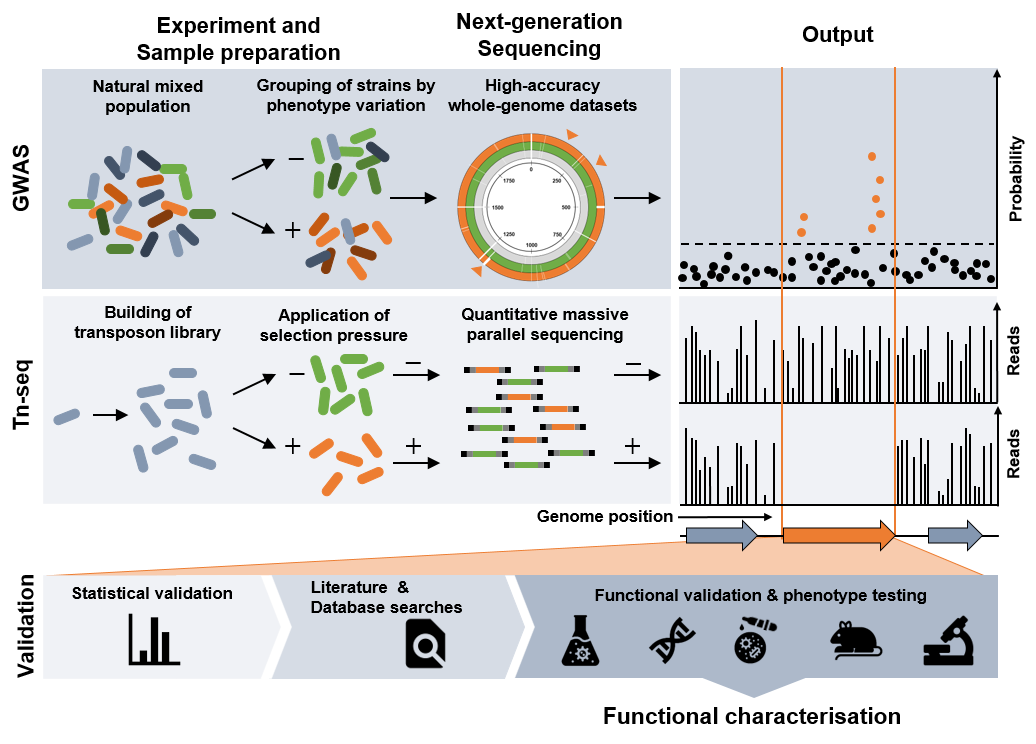
Schematic overview of population- and genome-wide screening methods discussed in the article, and a generalised validation pipeline that leads to functional characterisation of identified genetic variants.
In a joint interdisciplinary effort, researchers from the Florey Institute for Host-Pathogen Interactions at the University of Sheffield and the Milner Centre for Evolution at the University of Bath have now addressed this emerging schism in their latest review.
| Using their expertise bridging population and functional genomics with molecular microbiology, these researchers give their perspective of how to integrate new sequencing technologies and genomic screens with traditional microbiology. To this end, the researchers present a conceptual framework and practical roadmap for how to proceed from |
| comparative genomics to gene function. Furthermore, the article highlights key studies that already used an integrated approach to ‘next-generation microbiology’. | An integrated future for microbiology practice, outlined in this review article, should help microbiologists understand how to improve their research approaches. |
Prof. Samuel Sheppard, Professor and Director for Bioinformatics at the University of Bath said:
‘Population-wide genomic screens have revealed astounding genetic variation in bacteria. We must use this new information to learn more but we cannot ignore the scientific rigour of established molecular microbiology. With this article, we aim to provide a framework for integrating techniques for next generation microbiology.’
‘Population-wide genomic screens have revealed astounding genetic variation in bacteria. We must use this new information to learn more but we cannot ignore the scientific rigour of established molecular microbiology. With this article, we aim to provide a framework for integrating techniques for next generation microbiology.’
Dr Andrew Fenton, Lecturer at the University of Sheffield, added:
‘This review offers an interesting resource to molecular microbiologists and bioinformaticians alike; we hoped to write a thought-provoking piece which acts as a solid starting platform for early career researchers and students.’
‘This review offers an interesting resource to molecular microbiologists and bioinformaticians alike; we hoped to write a thought-provoking piece which acts as a solid starting platform for early career researchers and students.’
The article ‘Next-generation microbiology: from comparative genomics to gene function’ has recently been published in Genome Biology. Read the full article here.
This work was supported by grants from the Medical Research Council awarded to Prof. Samuel Sheppard and Dr Andrew Fenton.
This work was supported by grants from the Medical Research Council awarded to Prof. Samuel Sheppard and Dr Andrew Fenton.